How to Carry Out A Computerized Adaptive Test with Response time?
December 13, 2021
Process data tell a lot. It is a natural thought to utilize response time in computer-based testing since such data is readily available. Herein, I will illustrate how to incorporate response time in item selection methods for computerized adaptive testing. In this demo, you can see (a) the generation process for response time based on the lognormal model, (b) a complete routine for computerized adaptive testing, and (c) the performance of different item selection methods.
This demo for CAT with response time is based on the article: Choe, E. M., Kern, J. L., & Chang, H. H. (2018). Optimizing the use of response times for item selection in computerized adaptive testing. Journal of Educational and Behavioral Statistics, 43(2), 135-158.
Being interested in the Generalized MIT (GMIT) method, I tried to reproduce the results of Study 1 with set1 under the "v = 1, w = 0.5" condition. Three item selection methods were compared (i.e., Random selection method, maximum Fisher information selection method and GMIT method with v = 1, w = 0.5). It should be noticed that I did not implement any repetition to make it simple.
- Define function
- Set variables
I set some variables used in this study and generated item and person parameters in this part. - Main body
The estimation main body and the computation of seven evaluation criteria (i.e., bias and RMSE of ability, bias and RMSE of speed, M and SD of testing times, mean of testing overlap rate). - Save results
Save results and return evaluation indexs and timecost to the console.
Before we get started, it would be a good idea to clean our environment, record the starting time point, and set our working directory.
rm(list = ls())
time.start <- Sys.time()
# library packages ----------------------------------------
## This package can help you to produce samples from multivariate normal distribution
library(MASS)
# set working directory -----------------------------------
setwd("C:/Users/HYS/Desktop") ## You need to modify to your WDI introduced nine functions to implement the item selection methods, estimate examinees' abilities and speednesses, etc.
A. function1: itemInfo, compute Fisher information for dichotomous IRT model
B. function2: respon, generate response data for an examinee
C. function3: responTime, generate response time for an examinee
D. function4: selectRandom, Random selection method
E. function5: selectMFI, maximum Fisher information selection method
F. function6: selectGMIT, Generalized MIT method
G. function7: estMLE, estimate theta for an examinee with maximum likelihood method
H. function8: estEAP, estimate theta for an examinee with expected a posteriori method (as an interim substitute of MLE)
I. function9: estTau, estimate speed parameter for an examinee with maximum likelihood method
# define functions ----------------------------------------
itemInfo <- function(theta, a, b, c = 0, d = 1, D = 1.7){
# only for dichotomous IRT model
th <- theta
it <- cbind(a, b, c, d)
a <- it[, 1]
b <- it[, 2]
c <- it[, 3]
d <- it[, 4]
D <- D
e <- exp(D * a * (th - b))
Pi <- c + (d - c) * e/(1 + e)
Pi[Pi == 0] <- 1e-10
Pi[Pi == 1] <- 1 - 1e-10
dPi <- D * a * e * (d - c)/(1 + e)^2
P <- Pi
dP <- dPi
Q <- 1 - P
Ii <- dP^2/(P * Q)
}
respon <- function(theta, a, b, c = 0, D = 1.7){
## To generate response data for an examinee
ni <- length(a)
if (length(c) == 1) {
c <- rep(c, ni)
}
probs <- sapply(1:ni, function(i) {
c[i] + (1 - c[i])/(1 + exp(-D * a[i] * (theta - b[i])))
})
respondata <- ifelse(test = probs >= runif(n = ni, min = 0, max = 1), yes = 1, no = 0)
}
responTime <- function(tau, alpha, beta){
## To generate response time for an examinee
ni <- length(alpha)
time <- sapply(1:ni, function(i) {
rlnorm(n = 1, meanlog = beta[i] - tau, sdlog = 1/alpha[i])
})
}
selectRandom <- function(itemFlag){
## To select items randomly
nitemBank <- length(itemFlag)
rand <- sample(seq(1, sum(itemFlag)), 1)
id <- (1:nitemBank)[itemFlag > 0][rand]
}
selectMFI <- function(itemFlag, theta, a, b, c = 0, d = 1, D = 1.7){
## To select items according to the order of Fisher information at current estimated theta
info <- itemInfo(theta = theta, a = a, b = b, c = c, d = d, D = D)
ranks <- rank(info)
keepRank <- sort(ranks[itemFlag > 0], decreasing = TRUE)[1]
id <- which(ranks == keepRank & itemFlag == 1)
## items are selected randomly from those best ones
if (length(id) > 1){
id <- sample(id, 1)
}else{
id <- id
}
}
selectGMIT <- function(itemFlag, v, w, theta, tau, alpha, beta, a, b, c = 0, d = 1, D = 1.7){
## To select items with GMIT method
info <- itemInfo(theta = theta, a = a, b = b, c = c, d = d, D = D)
ET <- exp(beta - tau + 1/(2 * alpha^2))
deno <- abs(ET - v)^w
IT <- info/deno
ranks <- rank(IT)
keepRank <- sort(ranks[itemFlag > 0], decreasing = TRUE)[1]
id <- which(ranks == keepRank & itemFlag == 1)
## items are selected randomly from the those best ones
if (length(id) > 1){
id <- sample(id, 1)
}else{
id <- id
}
}
estMLE <- function (re, a, b, c = 0, D = 1.7, min = -4, max = 4){
## maximum likelihood estimates of theta for an examinee
## compute log-likelihood function
logf <- function(x, r, a, b, c, D) {
pi = c + (1 - c)/(1 + exp(-D * a * (x - b)))
ll = r * log(pi) + (1 - r) * log(1 - pi)
lf = sum(ll)
return(lf)
}
## find the optimal theta with max log-likelihood function
th <- optimize(logf, lower = min, upper = max, maximum = TRUE,
r = re, a = a, b = b, c = c, D = D)$maximum
return(th)
}
estEAP <- function(re, a, b, c = 0, D = 1.7, priorPar = c(0, 1), lower = -4, upper = 4, nqp = 40){
## expected a posteriori estimate of theta for an examinee
it <- cbind(a, b, c)
X <- seq(from = lower, to = upper, length = nqp)
## compute likelihood function
LL <- function(x, r, it, D) {
a <- it[, 1]
b <- it[, 2]
c <- it[, 3]
pi <- c + (1 - c)/(1 + exp(-D * a * (x - b)))
lls <- pi^r * (1 - pi)^(1 - r)
ll <- 1
for (i in 1:length(a)) ll <- ll * lls[i]
return(ll)
}
## use normal distribution as the prior distribution
num <- deno <- NULL
for (i in 1:length(X)) num[i] <- X[i] * dnorm(X[i], priorPar[1], priorPar[2]) * LL(X[i], re, it, D)
for (i in 1:length(X)) deno[i] <- dnorm(X[i], priorPar[1], priorPar[2]) * LL(X[i], re, it, D)
th <- sum(num)/sum(deno)
return(th)
}
estTau <- function(alpha, beta, logt){
## maximum likelihood estimates of speed for an examinee
num <- alpha^2 * (beta - logt)
deno <- alpha^2
speed <- sum(num)/sum(deno)
return(speed)
}Then, with the functions defined above, we can begin work on constructing a complete routine for CAT. To achieve this goal, we should first generate our examinees and the item bank, which are included in the "set variables" section. Later, a CAT with response time was presented in the "main body" section.
# set variables -------------------------------------------
nperson <- 1000
nitemBank <- 500
total.test.length <- 50
select.method <- c("Random", "MFI", "GMIT")
v <- 1
w <- 0.5
D <- 1.7
set.seed(13)
## generate item parameters
mu1 <- c(0.3, 0, 0)
sigma1 <- matrix(c(0.1, 0.15, 0,
0.15, 1, 0.25,
0, 0.25, 0.25), 3, 3)
itempara <- mvrnorm(n = nitemBank, mu1, sigma1)
a <- exp(itempara[, 1])
b <- itempara[, 2]
c <- rbeta(n = nitemBank, 2, 10)
alpha <- runif(n = nitemBank, min = 2, max = 4)
beta <- itempara[, 3]
## generate person parameters
mu2 <- c(0, 0)
sigma2 <- matrix(c(1, 0.25, 0.25, 0.25), 2, 2)
personpara <- mvrnorm(n = nperson, mu2, sigma2)
theta <- personpara[, 1]
tau <- personpara[, 2]
# main body -----------------------------------------------
estth.end <- esttau.end <- matrix(data = NA, nrow = nperson, ncol = length(select.method))
test.time <- matrix(data = NA, nrow = nperson, ncol = length(select.method))
ID.bank <- matrix(data = NA, nrow = nperson, ncol = total.test.length * length(select.method))
eval.index <- matrix(data = NA, nrow = 7, ncol = length(select.method))
for (m in 1:length(select.method)){
for (i in 1:nperson){
test.length <- 0
est.th <- est.tau <- 0
item.Flag <- rep(1, nitemBank)
response <- response.time <- test <- NULL
while(test.length < total.test.length) {
## randomly choose the first item
if (test.length == 0) {
item.ID <- selectRandom(itemFlag = item.Flag)
}else{
item.ID <- switch(m, selectRandom(item.Flag),
selectMFI(item.Flag, est.th, a, b, c),
selectGMIT(item.Flag, v, w, est.th, est.tau, alpha, beta, a, b, c))
}
item.Flag[item.ID] <- 0
test.length <- test.length + 1
## generate reponse socre and response time
response[test.length] <- respon(theta = theta[i], a = a[item.ID],
b = b[item.ID], c = c[item.ID])
response.time[test.length] <- responTime(tau = tau[i], alpha = alpha[item.ID],
beta = beta[item.ID])
test[test.length] <- item.ID
## estimate theta with MLE method (EAP as an interim substitute)
if(test.length >= 5 && sum(response) != 0 && sum(response) != test.length){
est.th <- estMLE(re = response, a = a[test], b = b[test], c = c[test], D = D)
}else{
est.th <- estEAP(re = response, a = a[test], b = b[test], c = c[test], D = D)
}
## estimate speed parameter with MLE method
est.tau <- estTau(alpha = alpha[test], beta = beta[test], logt = log(response.time))
}
estth.end[i, m] <- est.th
esttau.end[i, m] <- est.tau
test.time[i, m] <- sum(response.time)
## collect ID of selected items
ID.bank[i, (1 + total.test.length * (m - 1)) : (total.test.length * m)] <- test
## progress bar
print(paste("Finish: ", sprintf(fmt = '%0.1f', i/nperson * 100), "% for ", nperson,
" examinees with ", select.method[m], " item selection method.", sep = ""))
}
## compute evaluation indexs
eval.index[1, m] <- mean(theta - estth.end[, m]) # bias of theta
eval.index[2, m] <- sd(theta - estth.end[, m]) # RMSE of theta
eval.index[3, m] <- mean(tau - esttau.end[, m]) # bias of speed parameter
eval.index[4, m] <- sd(tau - esttau.end[, m]) # RMSE of speed parameter
eval.index[5, m] <- mean(test.time[, m]) # mean of testing times
eval.index[6, m] <- sd(test.time[, m]) # standard deviation of testing times
# compute the mean of testing overlap rate
item.overlap <- sapply(1:nitemBank, function(j){
used.times <- sum(ID.bank[, (1 + total.test.length * (m - 1)) : (total.test.length * m)] == j)
er <- used.times/nperson
})
eval.index[7, m] <- (nperson * sum(item.overlap^2)) / (total.test.length * (nperson - 1)) - 1/(nperson - 1)
}
The final part is to write your results. Let's see whether can you reach the same conclusion as Dr. Choe did. :D
# save results --------------------------------------------
## for theta
theta.result <- cbind.data.frame(estth.end, theta)
colnames(theta.result) <- c("Random", "MFI", "GMIT", "true_theta")
write.csv(x = theta.result, file = "est_theta.csv", row.names = FALSE)
## for speed parameter
tau.result <- cbind.data.frame(esttau.end, tau)
colnames(tau.result) <- c("Random", "MFI", "GMIT", "true_tau")
write.csv(x = tau.result, file = "est_tau.csv", row.names = FALSE)
## for test time
test.time.result <- test.time
colnames(test.time.result) <- c("Random", "MFI", "GMIT")
write.csv(x = test.time.result, file = "test_time.csv", row.names = FALSE)
## for evaluation indexs
eval.index.results <- eval.index
colnames(eval.index.results) <- c("Random", "MFI", "GMIT")
rownames(eval.index.results) <- c("bias_th", "RMSE_th", "bias_tau", "RMSE_tau",
"test_time_mean", "test_time_sd", "test_overlap_mean")
write.csv(x = eval.index.results, file = "evaluation_indexs.csv", row.names = TRUE)
timeCost <- Sys.time() - time.start
## return the results to the console
print(list(studyResults = eval.index.results, timeCost = timeCost))
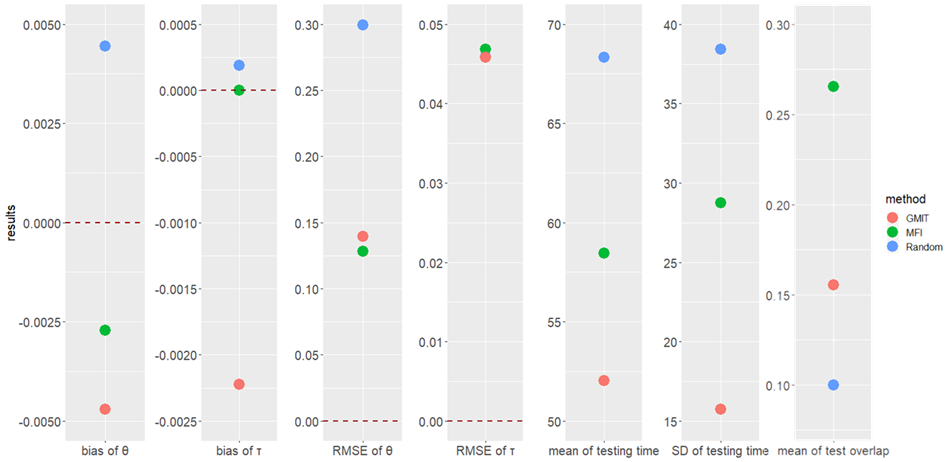
The scatter plot of the results.
Now you can use these codes to try using response times and build your own CAT!
Also, feel free to ask me any questions.
